This dataset is outdated, please check ToothFairy!
On March 30th, 2023 we released a new dataset named ToothFairy, which is an extension and improvement of this Maxillo dataset. We are also running a challenge on ToothFairy, so if you are interested please check it out. If you are trying to reproduce old experiments made on Maxillo, or you would like to compare a different network on the same kind of data you are free to go, otherwise there are no reasons to use this old version of the dataset.

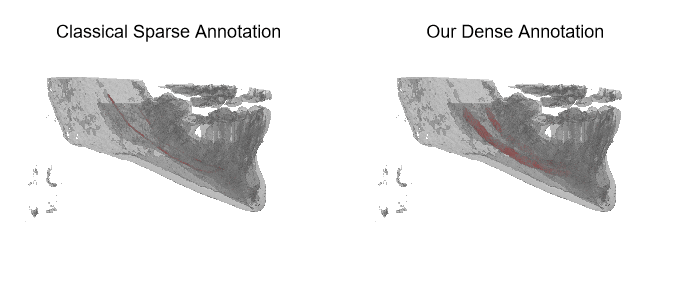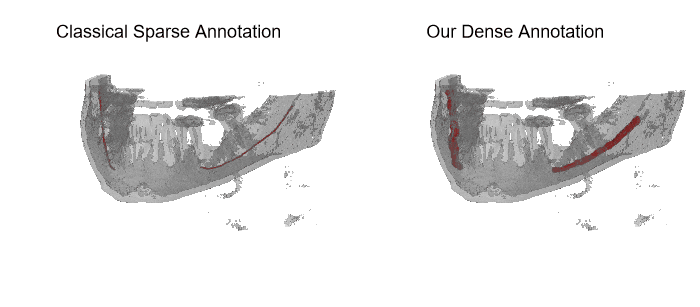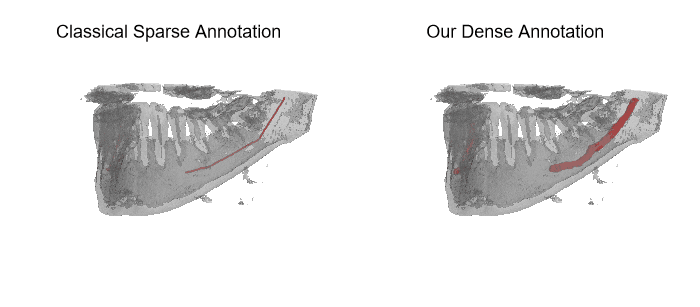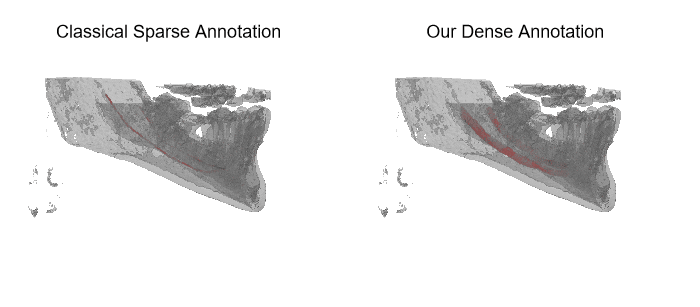
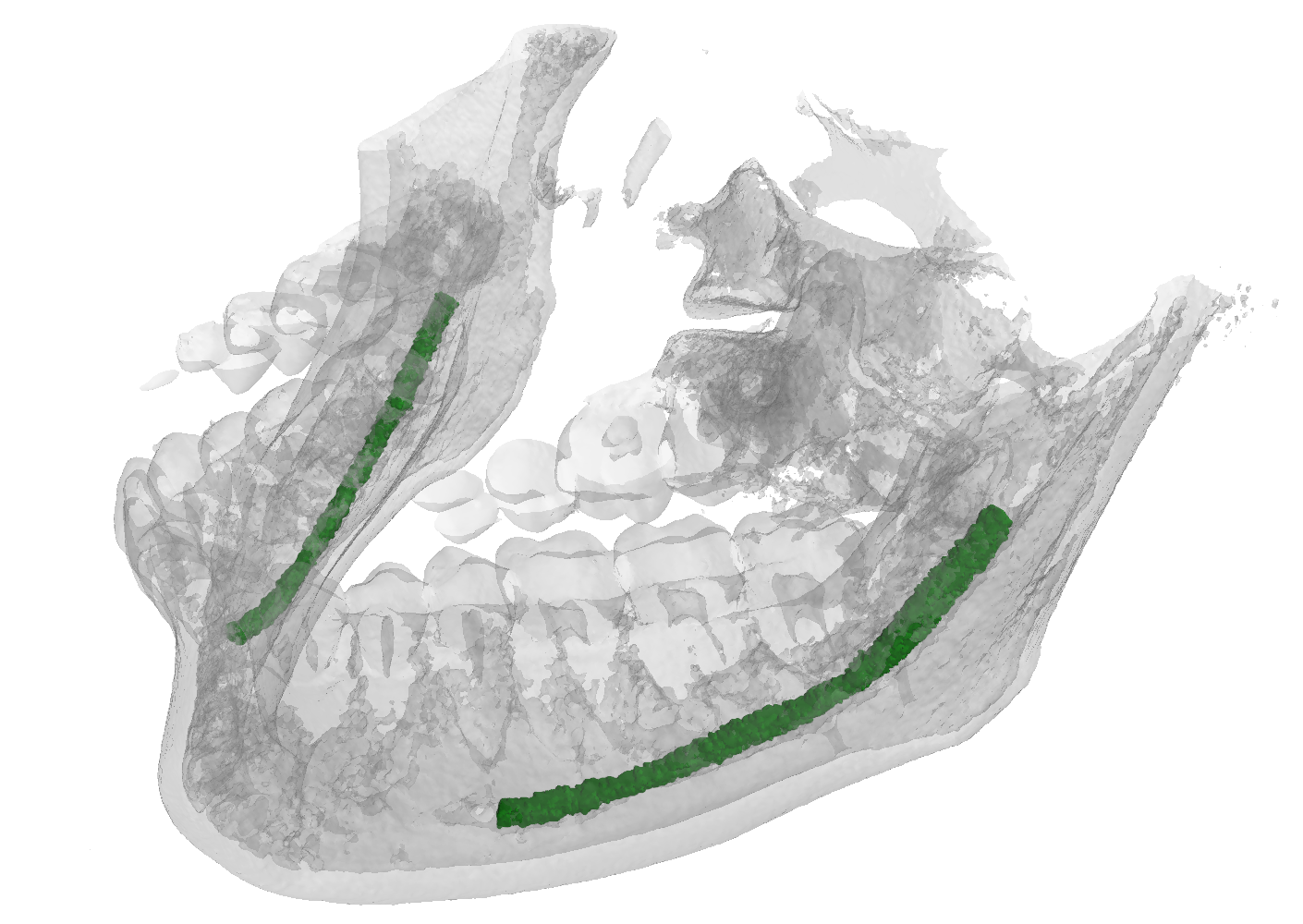
The IAN 3D dataset
Inferior Alveolar Nerve (IAN) canal detection has been the focus of multiple recent works in several branches of maxillo-facial imagery. Deep learning based techniques reached interesting results also in this research field, but the small size of 3D maxillofacial datasets strongly limited the performance of these algorithms. This forced researchers to build their own datasets, which are private, and this practice prevented the ability of really reproducing results and of fairly comparing proposals. We created a novel, large, and publicly available maxillo-facial CBCT (Cone Beam Computed Tomography) dataset, with 2D and 3D manual annotations, provided by expert clinicians. Leveraging on this dataset and employing deep learning techniques, we are able to improve the state of the art on the 3D mandibular canal segmentation. In this page you can download the data, the annotation tool and access to the source code which allows to exactly reproduce all the reported experiments.
Features
In the table below, we reported some basic information about our 3D images.
For a complete technical description of how we handled the volumes please refer to our manuscript.
Project is open source, feel free to contribute with new features.
| Field | Value |
|---|---|
| File format | Numpy |
| Values scale | Hounsfield |
| Avg volume shape | 169, 337, 381 |
| Max volume shape | 171, 423, 462 |
| Min volume shape | 148, 272, 334 |
| Primary dataset (dense) | 91 |
| Secondary dataset (sparse) | 256 |
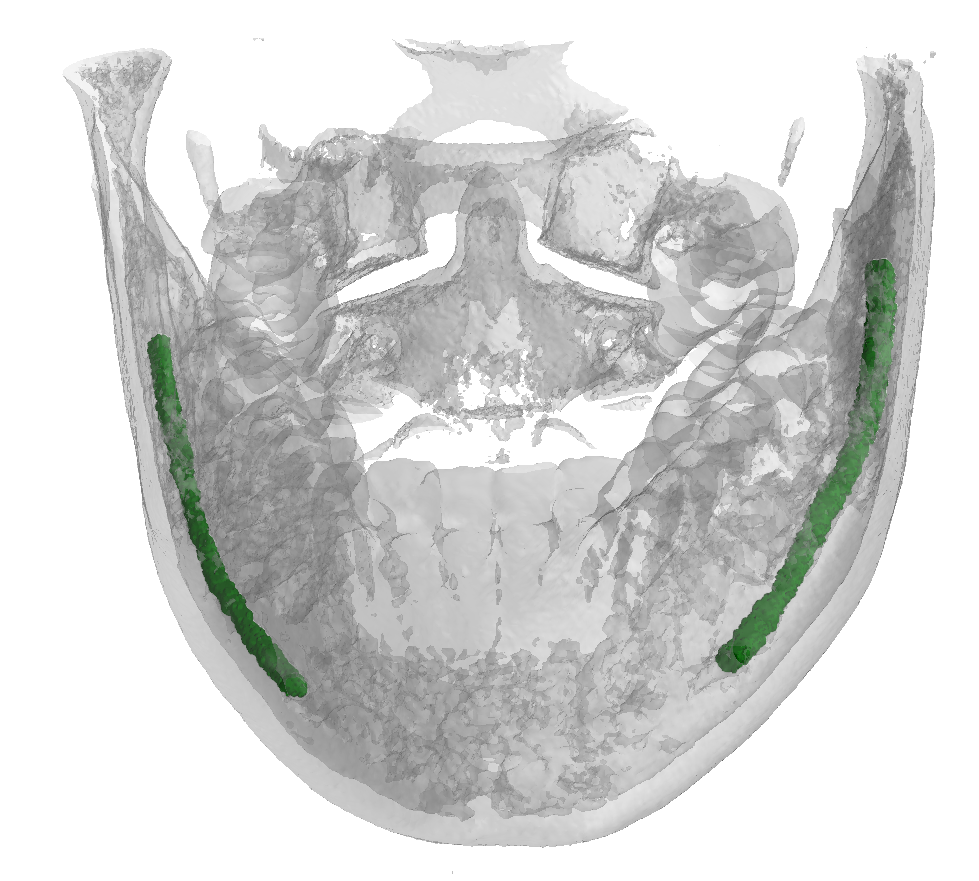
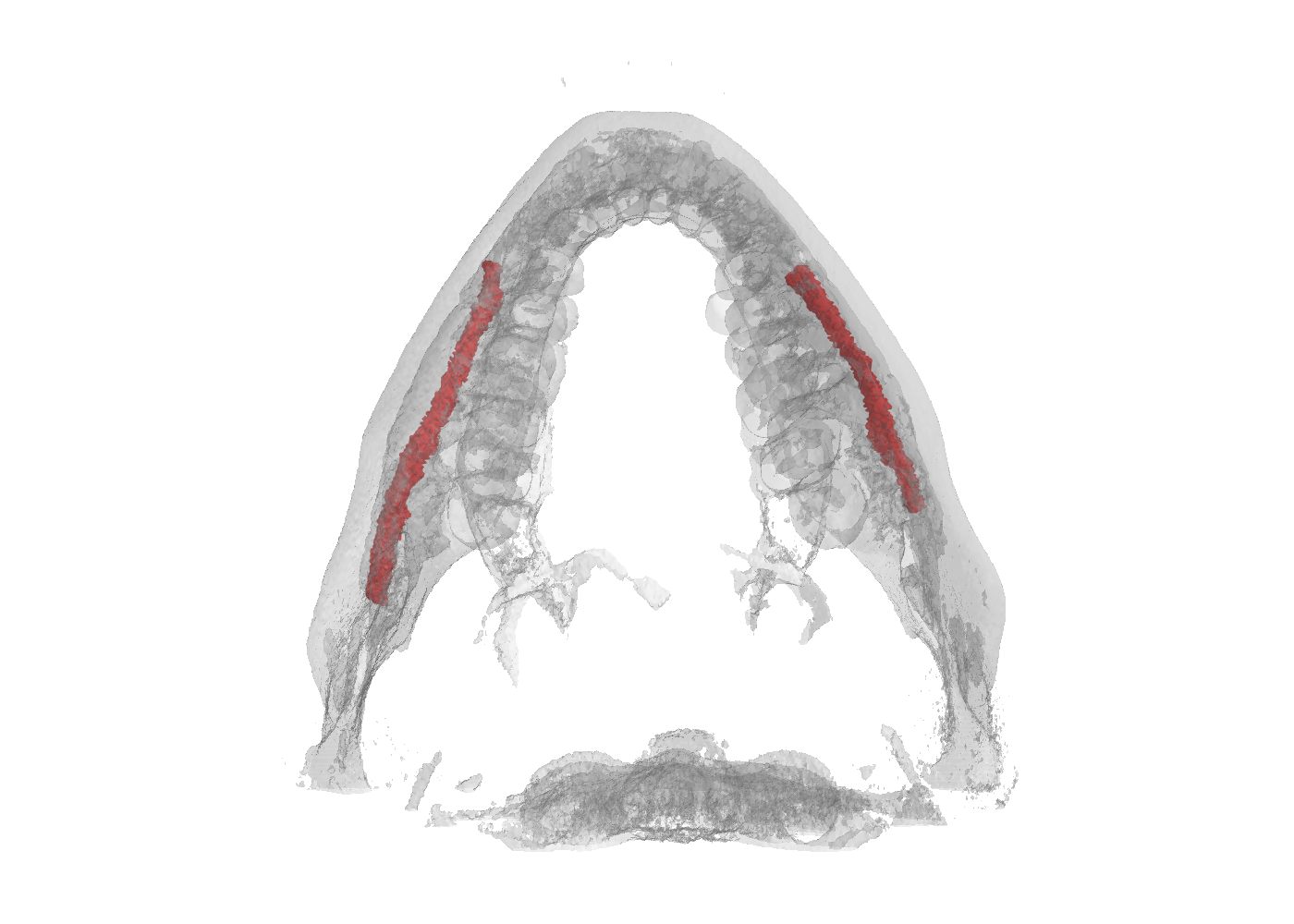
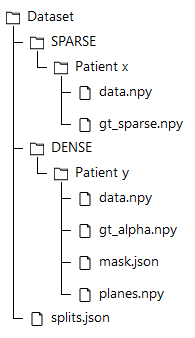
Dataset Tree
An overview of the folder's tree is pictured below. The split.json file provides information about which patients we used in our training, validation and test set. Each patient in the SPARSE folder contains a gt_sparse.npy numpy archive with the labels generated from the 2D panoramic view. Using the code provided in our repository, researchers can easily generate the circle expansion annoation from this file. A code to re-generate the gt_alpha.npy dense annotation from the masks.json and planes.npy files is also included in the repository of this project.
Download
Dataset is now available
Different resources are available for this project:
If you use our dataset, you must cite the following papers.
text loading...
Copy