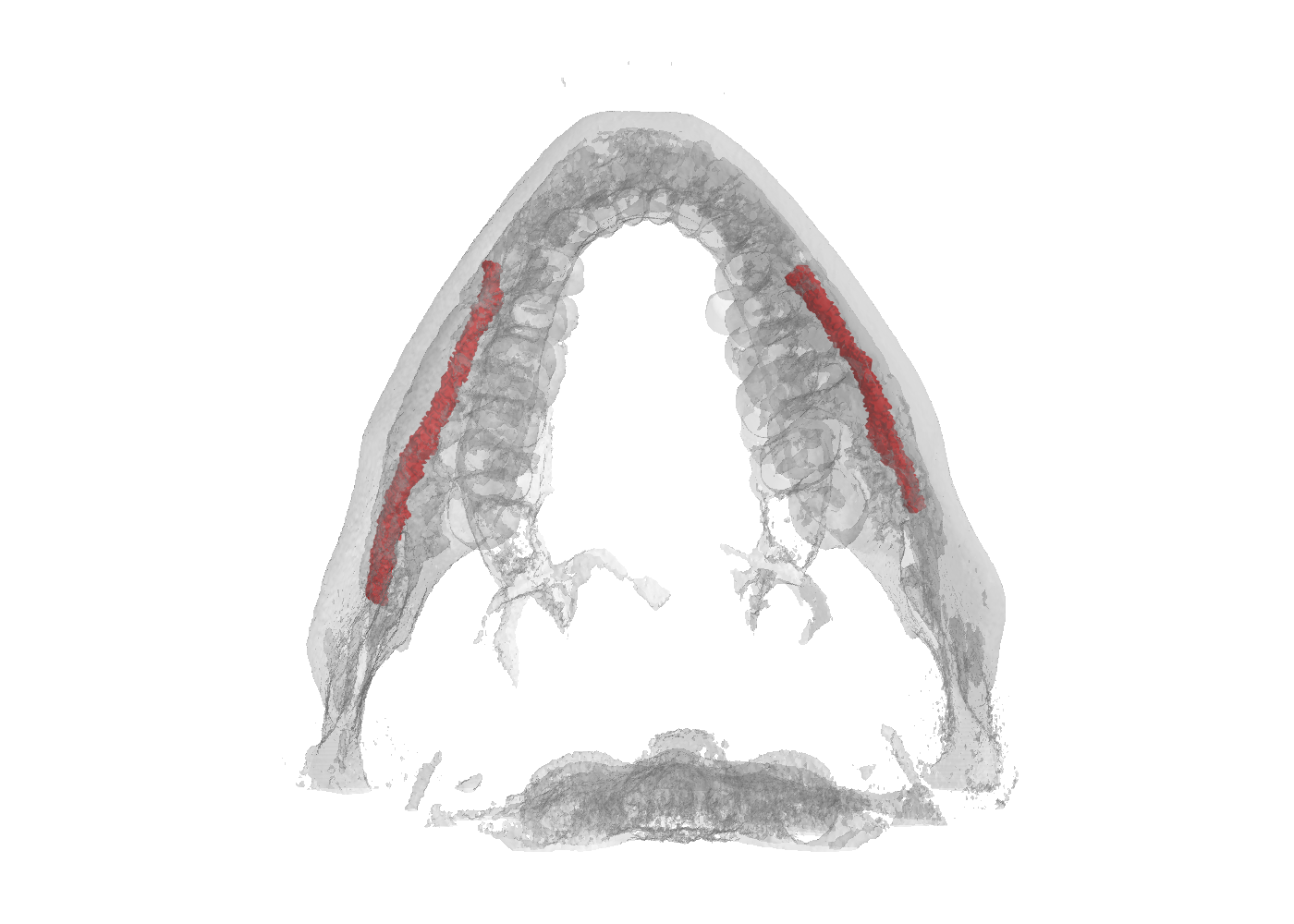
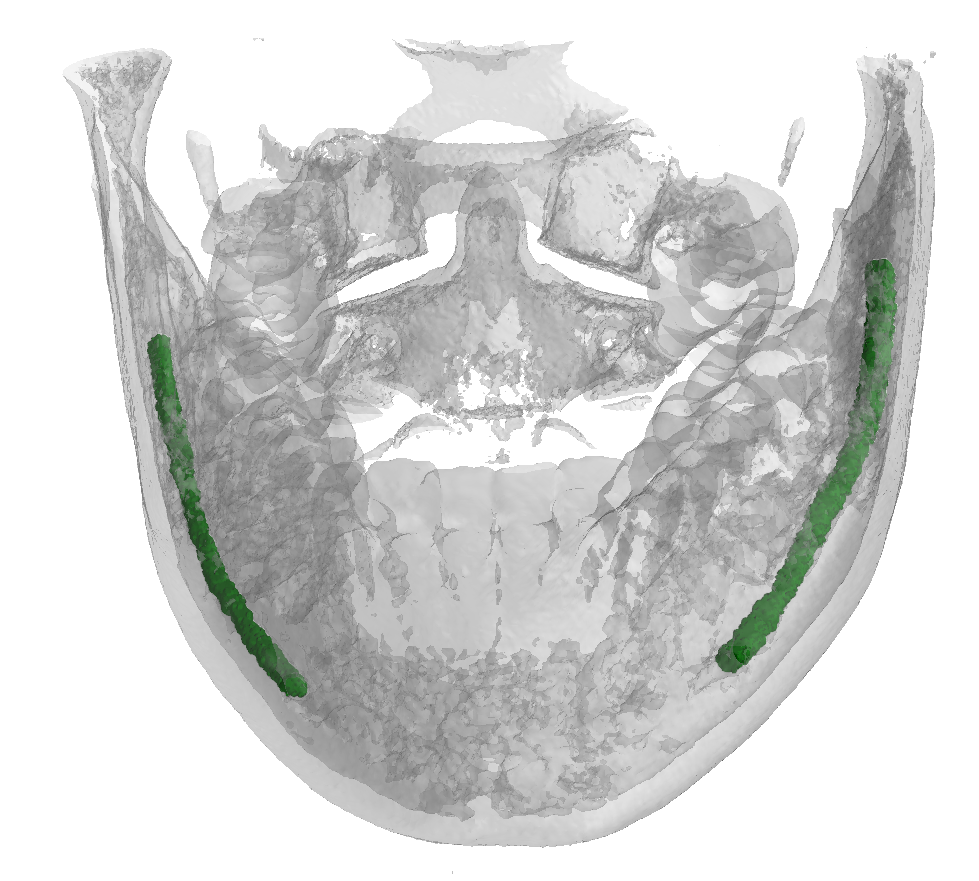
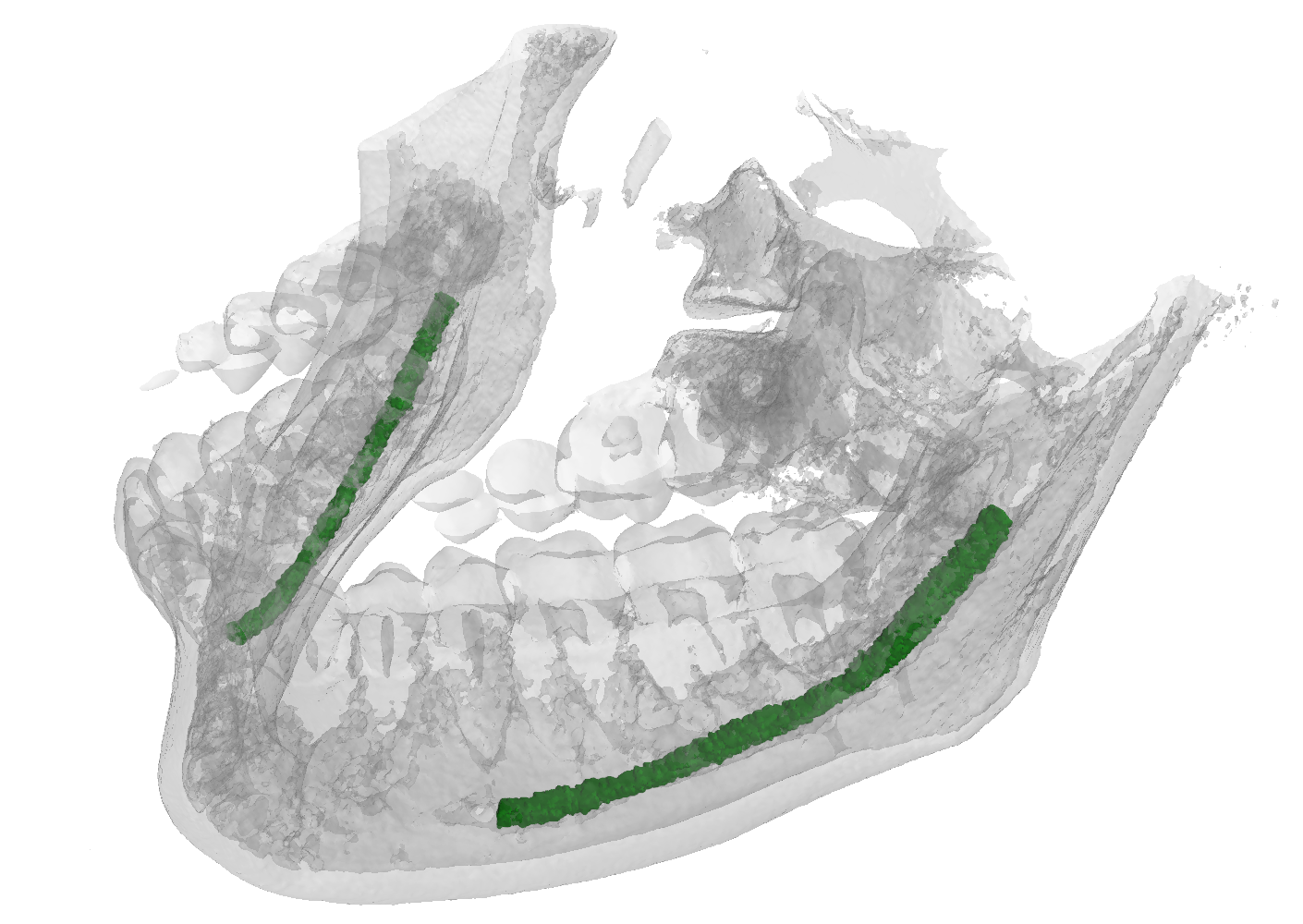
ToothFairy Dataset
This page describes the dataset associated to the ToothFairy challenge, a challenged organized by the University of Modena and Reggio Emilia with the collaboration of Radboud University. The challenge is hosted by grand-challenge and is part of MICCAI2023. For challenge details, please visit the dedicated website. The dataset used to run the challenge and here described is an extension and improvement of a previously released dataset.
Features
The table below resumes the core information about the (training) dataset released for MICCAI2023 ToothFairy challenge. For a complete technical description please refer to our CVPR paper and the structured challenge submission proposal.
| Field | Value |
|---|---|
| File Format | Numpy |
| Values Scale | Hounsfield |
| Avg Volume Shape | (169, 342, 370) |
| Max Volume Shape | (178, 423, 463) |
| Min Volume Shape | (148, 265, 312) |
| Secondary Dataset (only sparse annotations) | 290 (+34 w.r.t. the original Maxillo Dataset) |
| Primary Dataset (dense & sparse annotations) | 153 (+62 w.r.t. the original Maxillo Dataset) |
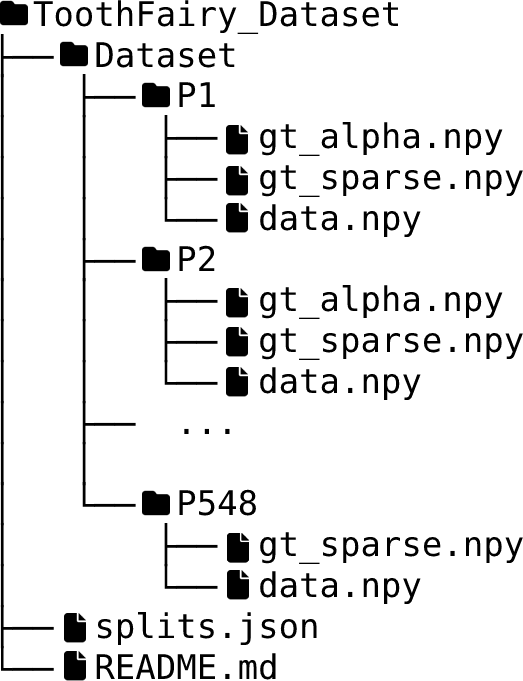
Dataset Tree
An overview of the folder's tree is pictured below. The splits.json file contains four different groups: training, validation, test, and synthetic. All the volumes inside the first three groups have both dense and sparse annotations, while the volumes within the synthetic group only have sparse annotations. We internally used this split to train the model provided as baseline for the challenge, please note that the official test set used in the challenge will not be publicly release. In other words, all patients have at least two files: data.npy, which corresponds to the CBCT scan, and gt_sparse.npy which is the sparse annotations (e.g., P548), some patients also have gt_alpha.npy which is the dense annotation (e.g. P1).
Download
Dataset is now available
The Download Dataset button will download the numpy dataset with both the sparse and dense annotation.
You can also download raw DICOM data (with only sparse annotations) by clicking the Download Raw Dataset button.
If you use our dataset, please cite our works in your manuscript.
text loading...
Copy