Visualizing Medical Image Data with Napari
When dealing with medical imaging, and more precisely with 3D volumes, specific tools are required to visualize them in an interactive way.
While there is a wide number of tools to visualize 3D data, the one we advice to adopt is Napari:
it is open-source, it perfectly integrates with Python, it supports barebone multiple helpful features, and allows us to develop plugins for it.
Load a ToothFairy Volume in Napari
We recommend taking a look at the official Napari website to know how to install and run it on your PC.
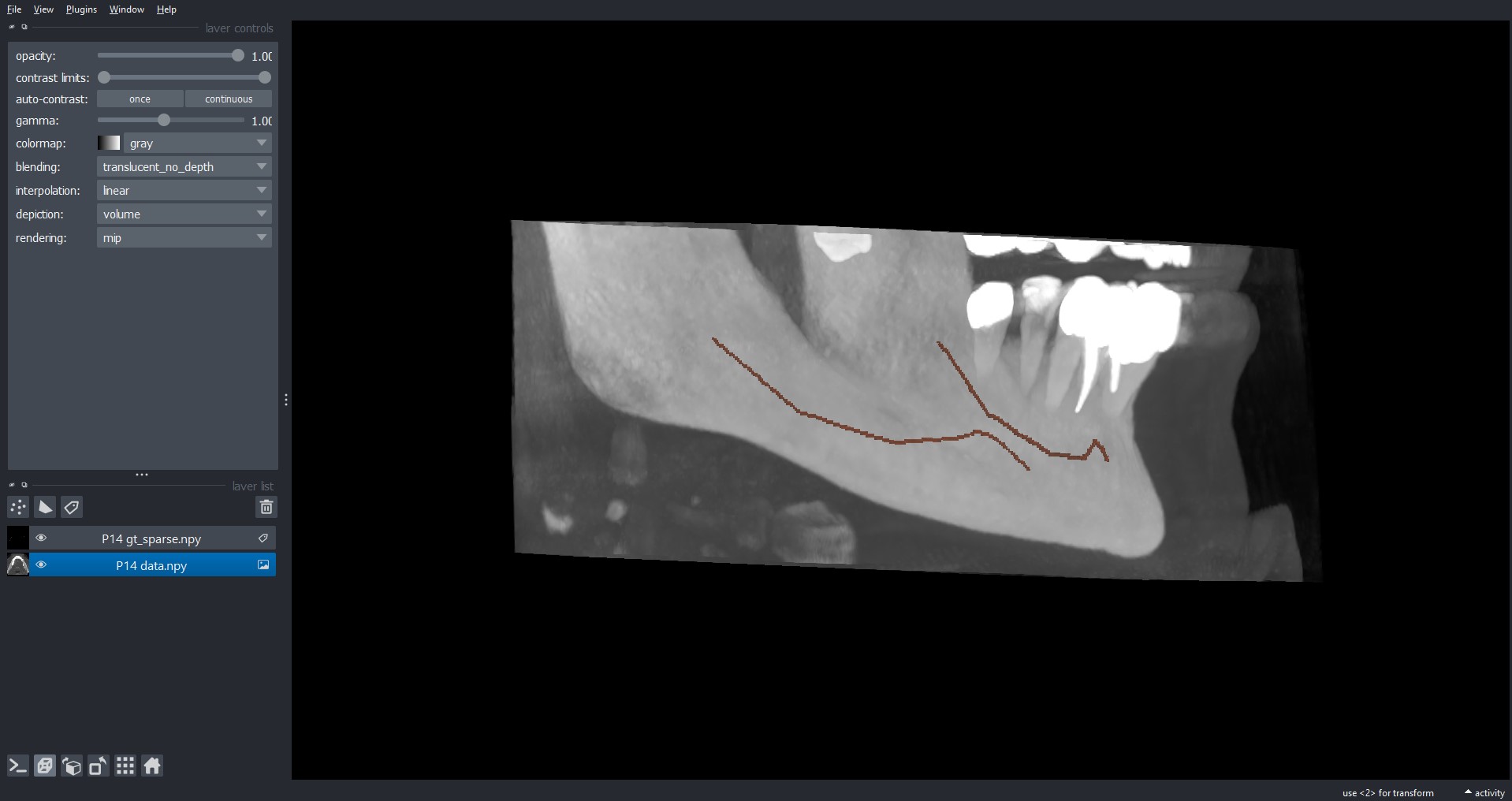
After installing, to load a volume from our dataset, you can simply click on File > Open File(s)... and
select the .npy file(s) you want to open.
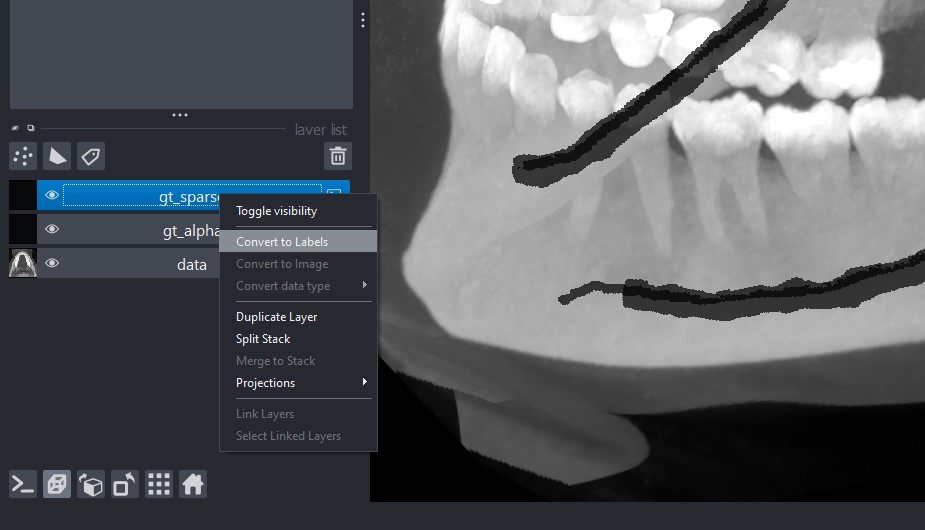
By default, Napari load the .npy file(s) as images, instead you prefer to look at gt_alpha.npy and
gt_sparse.npy as labels. To do so, after opening it, you can simply right-click on the specific layer
and select Convert to Labels.
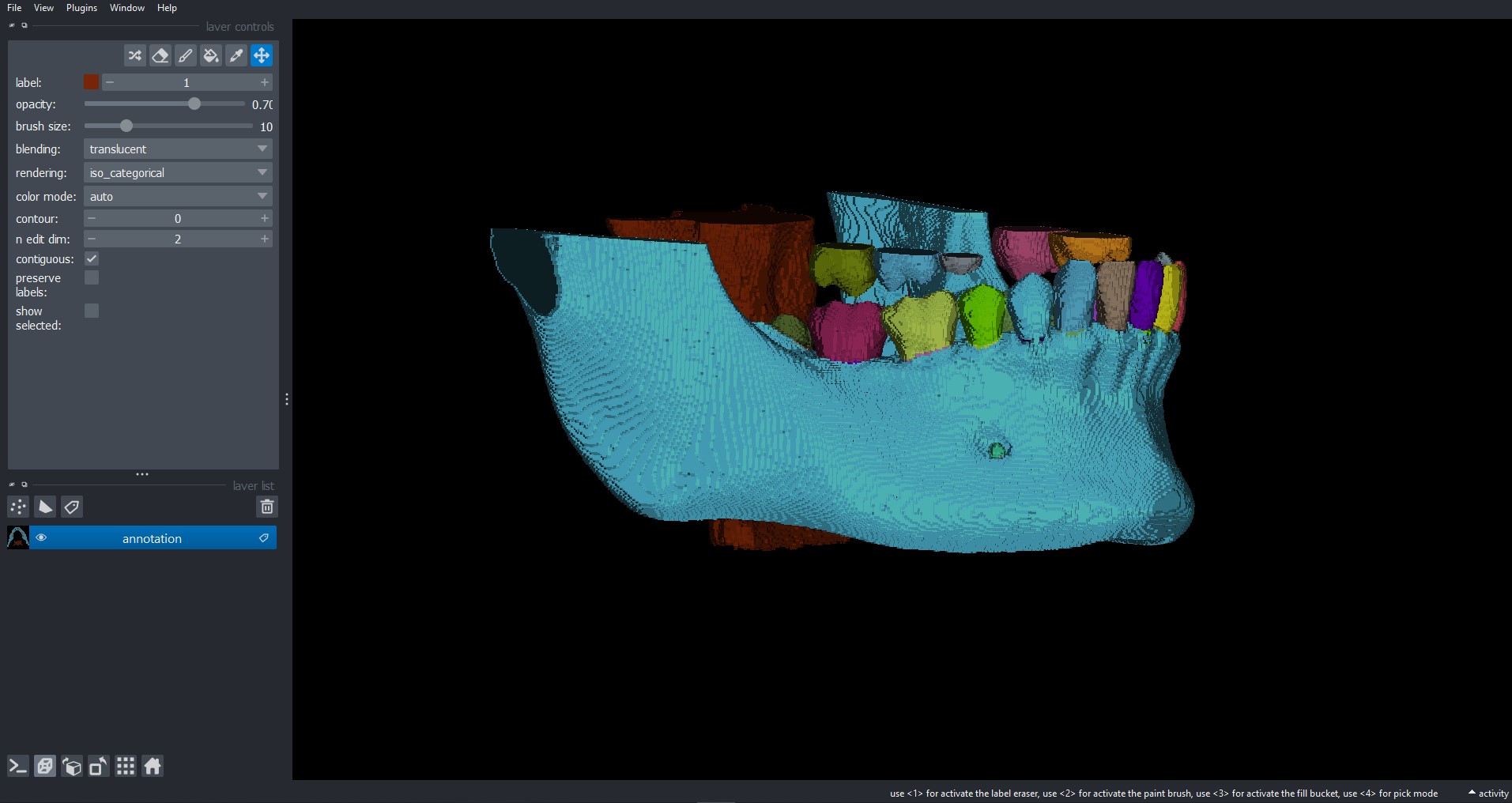
ToothFairy Napari Plugin
We are actively developing a specialized plugin that will enable users to visualize (and in the future) annotate these
3D numpy volumes from CBCT scans in the most comprehensive and user-friendly manner possible.
The plugin is freely available to the medical and research community through our dedicated
GitHub repository.